Chitinase Kinetics
A defined soluble substrate, 4-methylumbelliferyl ß-N,N',N''-triacetylchitotrioside, was used to characterize kinetic parameters of the barley chitinase. The pH profile shows activity is controlled by a base with a pKa of 3.9 (Glu 89) and an acid with a pKa of 6.9 (Glu 67) (Figure 1). The Km using the synthetic substrate is 33 µM, and the kcat is 0.33 min-1 while the Km for (GlcNAc)4 is 3 µM and kcat is 35 min-1(Figure 2A,B) . Binding constants were measured for ß-linked oligomers of N-acetylglucosamine. The monomer does not bind and dissociation constants for the dimer, trimer and tetramer are 43, 19, and 6 µM, respectively (Figure 3). Analysis of kinetic and dissociation constants proves the mechanism of barley chitinase is consistent with a Bi-Bi kinetic model for hydrolysis, with (GlcNAc)4 and water as substrates and (GlcNAc)2 as products (Figure 4). Substrate cleavage patterns show that (GlcNAc)6 is cleaved in half to (GlcNAc)3 as well as into (GlcNAc)4 and (GlcNAc)2 with almost equal efficiency. NMR analysis of cleavage products confirms that the enzyme proceeds with anomeric inversion of products.
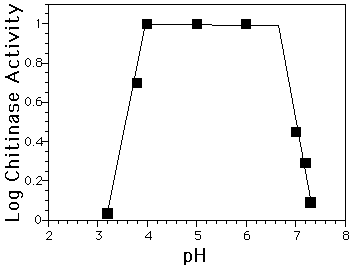
Figure 1: Log of chitinase activity vs pH. Inflection points give pKas of 3.8 and 6.8 which are presumably for Glu 89 and Glu 67, respectively
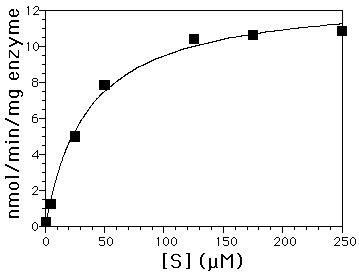
Figure 2A: Initial rate activity of barley chitinase. A direct plot of velocity data as a function of substrate concentrations shows hyperbolic kinetics. Vmax = 12 nmol/min/mg enzyme and Km = 33 µM for (GlcNAc)3UMB hydrolysis.
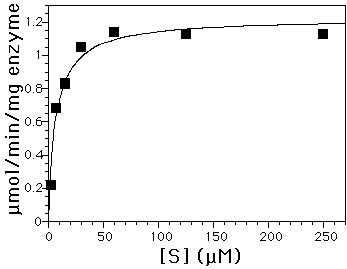
Figure 2B: Activity of barley chitinase with (GlcNAc)4. Vmax = 1.2 µmol/min/mg and Km = 3 µM
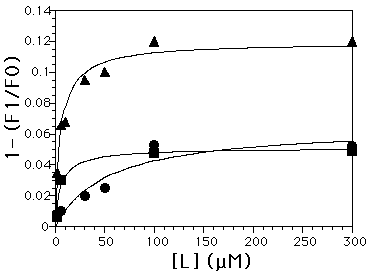
Figure 3: Chitinase binding of oligomers. F1/F0 = Fluorescence of chitinase with ligand/no ligand. A) triangles=(GlcNAc)4 (Kd = 6µM),
squares= (GlcNAc)3 (Kd = 19 µM), circles=(GlcNAc)2 (Kd = 43 µM). B) (GlcNAc)3UMB (Kd = 90 µM)
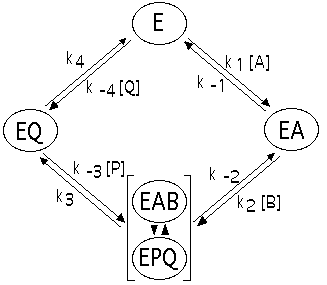
Figure 4: Kinetic model of chitinase hydrolysis. Reaction is treated as Bi Bi, with (GlcNAc)4 and H2O as the substrates; (GlcNAc)2 and (GlcNAc)2 as the products.




