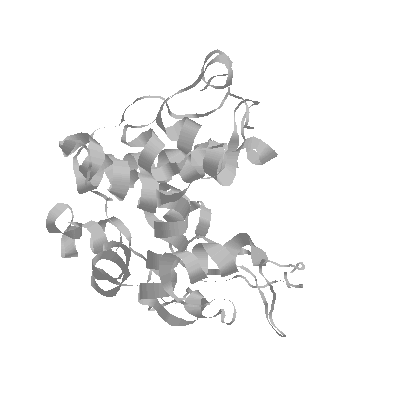
The X-ray structure of barley chitinase has been solved in our lab (Hart et al., 1993) and refined to 1.8 Å resolution (Hart et al., 1995). The structure shows a globular protein with high alpha-helical content and an elongated cleft running the length of the protein, presumably for substrate binding and catalysis. Hypothetical substrate binding models with the structure suggest that there are at least six sugar binding sites labeled A through F from the non-reducing end (Hart et al., 1995). Residues Glu 67 and Glu 89 are thought to be involved in hydrolytic catalysis, cleaving the substrate between sites D and E (Robertus et al., 1995). The putative mechanism involves protonation of the glycosidic bond at the C4 oxygen of the leaving E sugar, by Glu 67. At the same time, Glu 89 may act as a base to activate a water molecule which attacks the C1 position of the D sugar on the a side. This mechanism, suggesting that the barley chitinase catalysis proceeds with inversion of product, has been proven chemically.
You can view the structure of chitinase (and other proteins) by downloading the atomic coordinates (PDB file) from theBrookhaven PDB database. You will need a program such as RasMol to view the structure.
Hart, P.J., Monzingo, A.F., Ready, M.P., Ernst, S.R., & Robertus, J.D. (1993) Crystal Structure of an Endochitinase from Hordeum vulgare L. Seeds J. Mol. Biol. 229, 189-193.
Hart, P.J., Pfluger, H.D., Monzingo, A.F., Hollis, T., & Robertus, J.D. (1995) The Refined Crystal Structure of an Endochitinase from Hordeum vulgare L. Seeds at 1.8 A Resolution. J. Mol. Biol. 248, 402-413.
Robertus, J.D., Hart, P.J., Monzingo, A.F., Marcotte, E., & Hollis, T. (1995) The Structure of Chitinases and Prospects for Structure-Based Drug Design Can. J. Bot. 73, (Suppl. 1).
